About
Frequently Asked Questions
Brief introduction of development history of sgRNAcas9-AI. Potential applications of sgRNAcas9-AI, and the latest papers published by Xie's lab (used as a reference).
CRISPR Software R&D Platform (Development History)
2014
sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites
2017
CRISPR-offinder: a CRISPR sgRNA design and off-target searching tool for user-defined protospacer adjacent motif
2021
MagicEye: a moble Apps for CRISPR-based diagnostics technology
2022
sgRNAcas9-AI: optimized CRISPR sgRNA design for Cas9 variants by deep learning
Applications of sgRNAcas9-AI
CRISPR-based screening
CRISPR-based diagnostics
CRISPR-based Screening Platform
The successful adaptation of CRISPR–Cas9 approaches for genetic screens has become a powerful tool for the unbiased discovery of host dependent factor in mammalian cells. The sgRNAcas9-AI program can be used to quickly design optimized sgRNAs (or sgRNA library) for improved CRISPR screening efficiency.

(Citing: Zhang et al., J Genet Genomics, 2021)
1.Zhao C, Liu H, Xiao T, Wang Z, Nie X, Li X, Qian P, Qin L, Han X, Zhang J, Ruan J, Zhu M, Miao YL, Zuo B, Yang K, Xie S, Zhao S. CRISPR screening of porcine sgRNA library identifies host factors associated with Japanese encephalitis virus replication. Nat Commun. 2020 Oct 14;11(1):5178. doi: 10.1038/s41467-020-18936-1.
2.Sun L, Zhao C, Fu Z, Fu Y, Su Z, Li Y, Zhou Y, Tan Y, Li J, Xiang Y, Nie X, Zhang J, Liu F, Zhao S, Xie S, Peng G. Genome-scale CRISPR screen identifies TMEM41B as a multi-function host factor required for coronavirus replication. PLoS Pathog. 2021 Dec 6;17(12):e1010113. doi: 10.1371/journal.ppat.1010113. eCollection 2021 Dec.
CRISPR-based Diagnostics Platform
A field-deployable assay platform named RApid VIsual CRISPR (RAVI-CRISPR) based on a ROX-labeled reporter with isothermal amplification and CRISPR/Cas12a targeting was developed. The web version of sgRNAcas9-AI (Custom PAM) can be used to design crRNAs for CRISPR/Cas12, Cas13, etc.
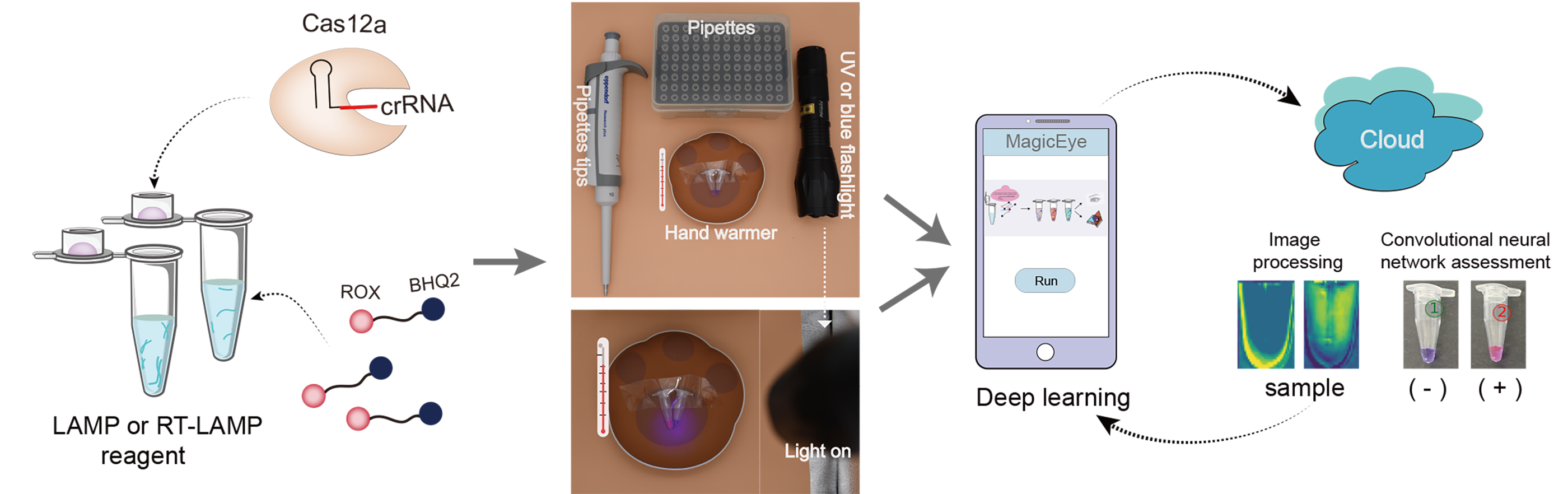
(Citing: Xie et al., ACS Synth Biol, 2022)
1.Xie S, Tao D, Fu Y, Xu B, Tang Y, Steinaa L, Hemmink JD, Pan W, Huang X, Nie X, Zhao C, Ruan J, Zhang Y, Han J, Fu L, Ma Y, Li X, Liu X, Zhao S. Rapid Visual CRISPR Assay: A Naked-Eye Colorimetric Detection Method for Nucleic Acids Based on CRISPR/Cas12a and a Convolutional Neural Network. ACS Synth Biol. 2022 Jan 21;11(1):383-396. doi: 10.1021/acssynbio.1c00474. Epub 2021 Dec 23.
2.Liu J, Tao D, Chen X, Shen L, Zhu L, Xu B, Liu H, Zhao S, Li X, Liu X, Xie S, Niu L. Detection of Four Porcine Enteric Coronaviruses Using CRISPR-Cas12a Combined with Multiplex Reverse Transcriptase Loop-Mediated Isothermal Amplification Assay. Viruses. 2022 Apr 17;14(4):833. doi: 10.3390/v14040833.
3.Liu S, Tao D, Liao Y, Yang Y, Sun S, Zhao Y, Yang P, Tang Y, Chen B, Liu Y, Xie S, Tang Z. Highly Sensitive CRISPR/Cas12a-Based Fluorescence Detection of Porcine Reproductive and Respiratory Syndrome Virus. ACS Synth Biol. 2021 Oct 15;10(10):2499-2507. doi: 10.1021/acssynbio.1c00103. Epub 2021 Sep 20.
4.Tao D, Liu J, Nie X, Xu B, Tran-Thi TN, Niu L, Liu X, Ruan J, Lan X, Peng G, Sun L, Ma Y, Li X, Li C, Zhao S, Xie S. Application of CRISPR-Cas12a Enhanced Fluorescence Assay Coupled with Nucleic Acid Amplification for the Sensitive Detection of African Swine Fever Virus. ACS Synth Biol. 2020 Sep 18;9(9):2339-2350. doi: 10.1021/acssynbio.0c00057. Epub 2020 Aug 31.
CRISPR technology
1.Han X, Gao Y, Li G, Xiong Y, Zhao C, Ruan J, Ma Y, Li X, Li C, Zhao S, Xie S. Enhancing the antibacterial activities of sow milk via site-specific knock-in of a lactoferrin gene in pigs using CRISPR/Cas9 technology. Cell Biosci. 2020 Nov 19;10(1):133. doi: 10.1186/s13578-020-00496-y.
2.Zhao C, Wang Y, Nie X, Han X, Liu H, Li G, Yang G, Ruan J, Ma Y, Li X, Cheng H, Zhao S, Fang Y, Xie S. Evaluation of the effects of sequence length and microsatellite instability on single-guide RNA activity and specificity. Int J Biol Sci. 2019 Oct 3;15(12):2641-2653. doi: 10.7150/ijbs.37152. eCollection 2019.
3.Zhang C, Zhou Y, Xie S, Yin Q, Tang C, Ni Z, Fei J, Zhang Y. CRISPR/Cas9-mediated genome editing reveals the synergistic effects of β-defensin family members on sperm maturation in rat epididymis. FASEB J. 2018 Mar;32(3):1354-1363. doi: 10.1096/fj.201700936R. Epub 2018 Jan 3.
4.Zhao C, Zheng X, Qu W, Li G, Li X, Miao YL, Han X, Liu X, Li Z, Ma Y, Shao Q, Li H, Sun F, Xie S, Zhao S. CRISPR-offinder: a CRISPR guide RNA design and off-target searching tool for user-defined protospacer adjacent motif. Int J Biol Sci. 2017 Nov 1;13(12):1470-1478. doi: 10.7150/ijbs.21312. eCollection 2017.
5.Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One. 2014 Jun 23;9(6):e100448. doi: 10.1371/journal.pone.0100448. eCollection 2014.
Systematic Review
1.Zhang J, Khazalwa EM, Abkallo HM, Zhou Y, Nie X, Ruan J, Zhao C, Wang J, Xu J, Li X, Zhao S, Zuo E, Steinaa L, Xie S. The advancements, challenges, and future implications of the CRISPR/Cas9 system in swine research. J Genet Genomics. 2021 May 20;48(5):347-360. doi: 10.1016/j.jgg.2021.03.015. Epub 2021 May 11.
2.Zhang D, Hussain A, Manghwar H, Xie K, Xie S, Zhao S, Larkin RM, Qing P, Jin S, Ding F. Genome editing with the CRISPR-Cas system: an art, ethics and global regulatory perspective. Plant Biotechnol J. 2020 Aug;18(8):1651-1669. doi: 10.1111/pbi.13383. Epub 2020 Apr 30.
3.Ahmad HI, Ahmad MJ, Asif AR, Adnan M, Iqbal MK, Mehmood K, Muhammad SA, Bhuiyan AA, Elokil A, Du X, Zhao C, Liu X, Xie S. A Review of CRISPR-Based Genome Editing: Survival, Evolution and Challenges. Curr Issues Mol Biol. 2018;28:47-68. doi: 10.21775/cimb.028.047. Epub 2018 Feb 11.